Troubleshooting
The search bar is disabled with the message “Initializing ImageJ…”
Since napari-imagej is calling Java code under the hood, it must launch a Java Virtual Machine (JVM). The JVM is not launched until the user starts napari-imagej. As we cannot search Java functionality until the JVM is running, the search bar is not enabled until the JVM is ready.
The first launch of napari-imagej can take significantly longer than subsequent launches while the underlying framework downloads the Java artifacts needed to run ImageJ2. Downloading these libraries can take minutes. These libraries are cached, however, so subsequent launches should not take more than a couple of seconds.
The image dimension labels are wrong in ImageJ after transferring from napari
Internally, napari does not utilize image dimension labels (i.e. X, Y, Channel, etc…) and instead assumes that the n-dimensional arrays (i.e images) conform to the scikit-image dimension order convention. ImageJ2 however does care about dimension labels and uses them to define certain operations.
For example, if you open the sample live cell wide-field microscopy data of dividing HeLa cell nuclei (which has the dimension order (X, Y, Time) in ImageJ’s convention) in napari, transfer the data over to ImageJ2 with the napari-imagej transfer button and examine the properties of the image you will find that ImageJ2 has confused the Time dimension for Channel. ImageJ2 thinks the transferred data has 40 channels instead of 40 frames.
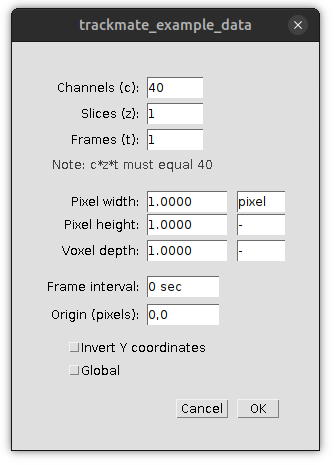
The reason this happens is because ImageJ2 is not given dimension labels when data is transferred from napari. When ImageJ2 has no dimension label information for a given image then the (X, Y, Channel, Z, Time) dimension order and labels are applied to the image. In this example, the X and Y dimension labels are set properly, but the last dimension (which we know should be Time) is set to Channel. Note that this also means if your napari image has a shape that does conform to the scikit-image dimension order (t, pln, row, col, ch) it is possible that transferred images could be transposed into unintended orthogonal views of the data.
To fix the dimension labels on your image data open the image’s properties and assign the correct dimension value to the appropriate field. In this example we want to assign Channel (c) the value 1 (there is only 1 channel) and Frames (t) the value 40 (there are 40 frames in the dataset). You can also set the unit type (e.g micron, pixel, etc…) and size for the image in the image properties.